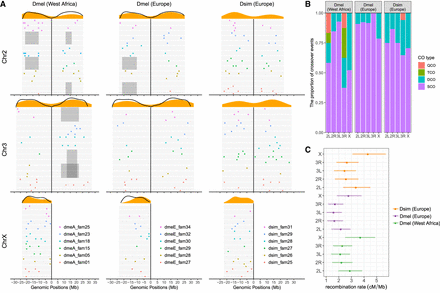
A summary of recombination (crossovers) in the females of three populations of Drosophila. (A) The genomic positions of breakpoints on chromosomes. Each gray bar represents an offspring individual and colors denote the breakpoints in different families. Orange density plots above represent the density of breakpoint along each chromosome and the black curve lines represent recombination rates from Comeron et al. (2012). The classical inversions of D. melanogaster (In(2L)t, In(2R)NS, In(3R)K, and In(3R)Payne) heterozygous in the parents are shown as gray areas. (B) The proportion of crossover types; SCO: one crossover event on a chromosomal arm during one meiotic process; DCO: two crossover events in one meiotic process; TCO: three crossover events; QCO: four crossover events. (C) The recombination rate and 95% CI estimated from a Bayesian GLMM in which “population” and “chromosome” were fitted as fixed effects and parental ID as a random effect, but with no effect of inversion fitted (see the main text for alternative estimates).











