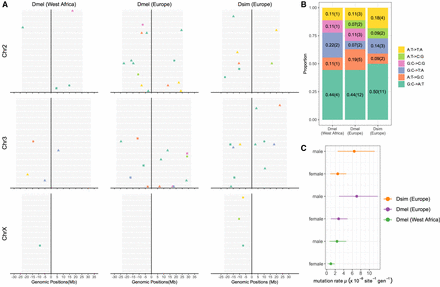
A summary of de novo SNMs identified in a West African and a European population of Drosophila melanogaster, and a European population of Drosophila simulans. (A) The genomic positions of the SNMs on chromosomes. Each gray bar represents the genome of one offspring. Point color represents mutation type and point shape represents the parental origin of the mutation (triangle: paternal; circle: maternal). Square points are used to denote the SNMs with unknown parental origin caused by the lack of informative surrounding SNP markers. (B) The SNM spectrum. The y-axis shows the proportion of each mutation type, and the numbers in brackets are the counts. (C) The mutation rate of SNMs and 95% CI estimated from our main Bayesian GLMM in which “population” and “sex” were fixed effects and parental ID was a random effect.











