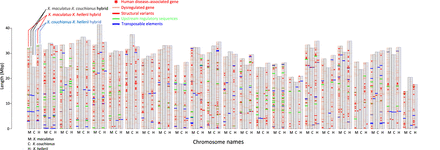
Chromosomal distribution of dysregulated genes in reciprocal Xiphophorus hybrids. Chromosomal distribution of hybrid dysregulated genes and their association to structural variants, transposable elements, and upstream regulatory sequences in the parental genomes. (M) X. maculatus; (C) X. couchianus; (H) X. hellerii. Reciprocal F1 interspecies hybrids are produced between X. maculatus, X. couchianus, and X. hellerii. Dysregulation of the genes is identified by comparing hybrid gene expression to parental species and determined if their expression pattern is different from the parentals (i.e., transgressively expressed in hybrid). A clustered bar graph is used to show the genomic locations of dysregulated genes. Bar height represents chromosome length. Because each species is involved in two types of hybrids, each chromosome per species is split in the middle, with the left and right halves representing chromosome in the two types of hybrids, as illustrated in the figure. Pink bars represent the chromosomal location of dysregulated genes, with red, green, and blue bars highlighting dysregulated genes adjacent to inter-specific structural variant(s) polymorphisms or showing polymorphisms in upstream regulatory sequences or transposable elements in their −1000 to zero of transcription start site. If the human ortholog of a Xiphophorus dysregulated gene is known to be related to disease, the Xiphophorus gene is labeled with an asterisk.











