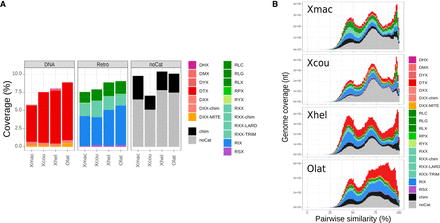
TE analyses in Xiphophorus. (Xmac) X. maculatus; (Xcou) X. couchianus; (Xhel) X. hellerii; (Olat) Oryzias latipes. (A) TE coverages are similar between the surveyed species, and TEs are equally distributed between class II (DNA transposons) and class I (retrotransposons). (DHX) Helitron; (DMX) Maverick; (DYX) Crypton; (DTX) TIR transposon; (DXX) unclassified DNA element; (chim) chimeric element; (RLC) Copia; (RLG) Gypsy; (RLX) LTR retrotransposon; (RPX) Penelope; (RYX) DIRS; (RXX) unclassified retrotransposon; (RIX) LINE; (RSX) SINE. (B) TE landscapes in Xiphophorus species and medaka species. TE insertions were compared between each other for each identified TE family; the distribution of similarity scores was computed and converted into genomic coverage according to the coverage of the TE family. The right extremity of the graph corresponds to recent (highly similar) TE insertions.











