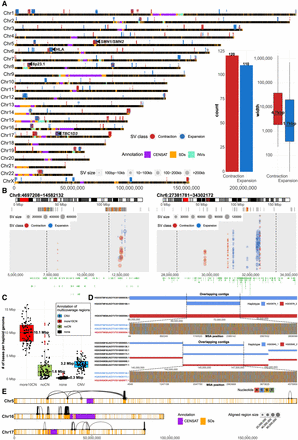
Tracking contig alignment discontinuities and multicoverage regions. (A) Genome-wide distribution of frequent (n = 230) contig alignment discontinuities (1 kbp to 1 Mbp in size). Each gap is represented in each separate assembly (HPRC, 94; HGSVC, 28) by a colored dot (blue, expansion [INS]; red, contraction [DEL]), and the size of each dot represents the size of the event in contig coordinates. A region is defined as an INS (blue) if there is a gap in a contig alignment (in reference T2T-CHM13, v1.1 coordinates) that is smaller than the sequence within a contig itself delineated by the left and right alignments flanking the gap. In contrast, a DEL (red) is defined as a gap in a contig alignment (in reference T2T-CHM13, v1.1 coordinates) that is larger than the sequence within a contig itself delineated by the left and right alignments around the gap. Putative expansions and contractions above the horizontal chromosomal lines were detected in HPRC assemblies, and those below the lines in HGSVC assemblies. Centromeric satellite regions are highlighted by gray rectangles and regions of segmental duplications (SDs) as orange rectangles on top of each chromosomal line (black). (B) Example regions (left, defensin locus, 8p23.1; right, HLA locus) with frequent expansions and contractions. Each region is highlighted as a red rectangle on chromosome-specific ideogram (top track). Below, there is an SD annotation for a given region represented as a set of rectangles colored by sequence identity. Expansions and contractions of each contig alignment with respect to the reference (T2T-CHM13, v1.1) are depicted as blue and red dots, respectively. The size of each dot represents the size of an event. (C) Assignment of total number base pairs covered by multiple contig alignments, in each haploid genome (n = 88), into four categories based on agreement with short-read-based CNV profiles (for detailed description of categories, see Methods). (D) Example regions in samples HG03579 and HG03540, where overlapping contigs associate with loss of heterozygosity. Top track shows contig alignments in a given region separately for haplotype 1 (blue; paternal) and haplotype 2 (red; maternal). Overlapping contig alignments are stacked on top of each other. The bottom track shows all variable positions detected in a multiple sequence alignment (MSA) over the region where contigs overlap (dashed lines). Here, one of the paternal contigs is nearly identical to a maternal contig at the region where contigs overlap. (E) Chromosomes 5, 16, and 17 are depicted as horizontal bars with the locations of SDs and centromeric regions highlighted as orange and purple rectangles, respectively. Contig alignment ends divided into multiple pieces are visualized as links between subsequent pieces of a single contig aligned to the reference (T2T-CHM13 v1.1). The length of the aligned pieces of a contig are defined by the size of each dot.











