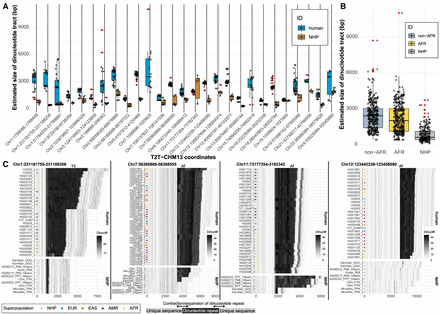
Sequence variation in low-complexity regions. (A) Size distribution comparison of dinucleotide tracts (y-axis) between human (blue) and nonhuman primates (NHPs; brown) for 27 selected regions (Methods). Outliers are highlighted as red dots. (B) A summary of size distribution of dinucleotide tracts (y-axis) between human samples of African (AFR; yellow) and non-African (non-AFR; light blue) origin and NHPs (gray) across all complete assemblies from 27 selected regions. (C) Difference in dinucleotide frequency (TC, AT) between humans and NHP in four genomic regions. Shades of gray color reflect the number of detected dinucleotides (defined at the top of each plot) in 100-bp-long DNA sequence chunks. Assembly names (y-axis) from NHP contain sample IDs and species-specific ID: (PTR) Pan troglodytes, (GGO) Gorilla gorilla, (PPA) Pan paniscus, (MMU) Macaca mulatta, (PAB) Pongo abelii, (PPY) Pongo pygmaeus. Numbers 1 and 2 represent parental homolog IDs of given sample assembly.











