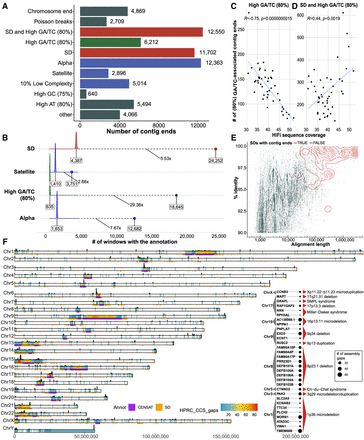
Sequence properties at defined contig ends. (A) The number of simple contig ends that are within or near (at most 10 kbp) a particular sequence annotation. Annotations are nonredundant and are prioritized in the order shown; for example, if a contig end is near the end of a chromosome and in an SD, it will only be annotated as a chromosome end. Note that chromosome ends are contig ends within the last 100 kbp of contigs. Poisson ends are contig ends that happen in only one haplotype (nonrecurrent and therefore likely to be random). SD and high GA/TC mean that the end is within 10 kbp of an SD and within 10 kbp of a 1-kbp window with at least 80% GA/TC content. (B) The fold enrichment in the number of contigs ends within 10 kbp of a sequence annotation compared with a distribution of randomly placed contig end simulations (10,000 permutations). Shown in text is the median of the random distribution (left), the fold enrichment (middle), and the observed value (right). In this analysis contig ends may exist in multiple categories; for example, if a contig end is near both an SD and a satellite sequence, it will appear in both simulations. (C) The effect of HiFi coverage on number of GA/TC breaks is negatively correlated when considered independently; however, when combined with SDs, the trend is inverted, as shown in D. (E) All SDs in T2T-CHM13 displayed by their length and percentage of identity (blue) versus the SDs that intersect contig ends (red). (F) Genome-wide distribution of gaps defined in between contig alignment ends (Methods) across all HPRC assemblies (n = 94). Color range reflects the number of assembly gaps overlapping each other in any given genomic region. On the top of each chromosomal bar, there is a density of simple contig ends. The height of each bar reflects the number of simple contig ends counted in 200-kbp-long genomic bins. Inset: List of protein-coding genes (n = 31) overlapping assembly breaks and reported microdeletion and microduplication syndromes.











