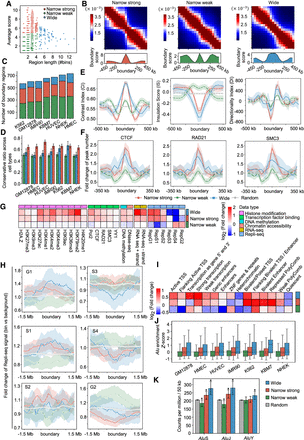
Three types of boundary regions defined based on the TAD separation landscape. (A) Scatter plot of boundary regions based on their lengths and average boundary scores. (B) Aggregated Hi-C contact maps around the NSBs (left), NWBs (middle), and WBs (right), combined with the average boundary score profiles in GM12878. (C) Number of three types of boundary regions identified in seven cell lines. (D) Conservation of three types of boundary regions among the seven cell lines. (E) Profiles of 1D indicators including CI (left), IS (middle), and DI (right) around different types of boundaries and random control in GM12878. The shaded areas represent the 95% confidence intervals in 1000 bootstraps. (F) Profiles of peaks fold change of TFs including CTCF (left), RAD21 (middle), and SMC3 (right) around different types of boundaries and randomly selected regions in GM12878. The fold changes are calculated by dividing the peak numbers in each bin by the average peak numbers across the chromosome. (G) Fold change profiles of multiple types of biological data constructed for three types of boundary regions in GM12878. Fold change of a biological marker in each type is defined as the median signal in boundary regions divided by the median signal across the chromosome. (H) Fold change profiles of Repli-seq signal around three types of boundaries and randomly selected regions in six different phases of the cell cycle: G1, S1, S2, S3, S4, and G2. (I) Fold change profiles of ChromHMM states constructed for three types of boundary regions in GM12878. The fold change of a chromatin state in each type is defined as the total length of the state in boundary regions divided by the expected state length in chromosome background. (J) Enrichment of Alu elements in three types of boundaries for seven cell lines. (K) Density of the Alu subfamilies in three types of boundaries and randomly selected regions in GM12878. Mann–Whitney U tests were performed between boundary regions and random regions, respectively. (*) P-value < 0.01.











