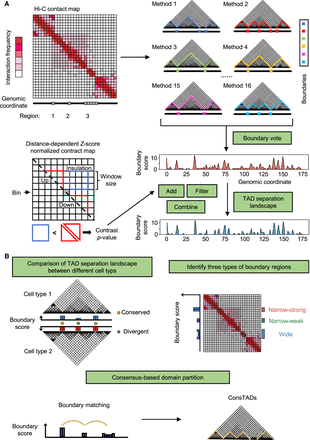
Overview of the construction of the TAD separation landscape and its three applications. (A) The representative Hi-C contact map contains several candidate boundary regions, and different TAD-calling methods identify diverse TADs with variable boundaries. Based on the boundary voting strategy, each bin on the genome will get a score to indicate how many methods consider it to be a TAD boundary. The TAD separation landscape is constructed from the original boundary score profile using three additional operations, including add, filter, and combine, which are calculated based on the contrast P-value, an indicator to reflect the insulation effect of chromatin interactions for each bin. (B) Three applications of the TAD separation landscape, including domain boundary comparison, boundary type identification, and consensus domain detection.











