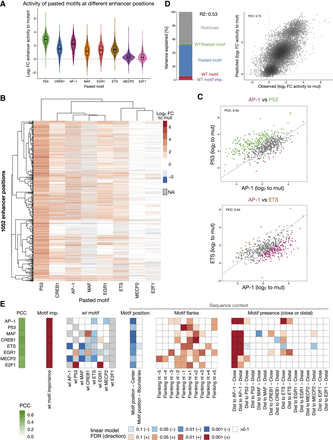
Human TF motifs require specific enhancer sequence contexts. (A) Distribution of enhancer activity changes (log2 FC to mutated sequence) across all enhancer positions for each pasted TF motif. (B) Heatmap of enhancer activity changes (log2 FC to mutated sequence) after pasting each of the eight selected human TF motifs in 1052 enhancer positions (positions with data for at least six motifs). TF motifs and positions were clustered using hierarchical clustering and the activity is colored in shades of red (activating) and blue (repressing); missing values are colored in gray. (C) Human TF motifs work differently at different enhancer positions. Comparison between enhancer activity changes (log2 FC to mutated sequence) after pasting AP-1 (x-axis) and P53 (top) or ETS (bottom) (y-axis), across all enhancer positions. Positions with stronger activity of each motif (≥twofold in respect to the other motif in the scatter plot) are colored (P53: green, AP-1: purple, ETS: brown). PCC: Pearson correlation coefficient. (D) TF motif activity in function of wild-type and pasted motif identity. Left: Bar plot showing the amount of variance explained by the wild-type motif importance and identity, the pasted motif identity, and the interaction between the wild-type and pasted motifs, using a linear model fit on all motif pasting results. Right: Scatter plots of predicted (linear model) versus observed enhancer activity changes (log2 FC to mutated sequence) across all motif pasting experiments. Color reflects point density. (E) Motif syntax rules modulate the function of human TF motifs. For each TF motif type (rows), we built a linear model to predict their activity across all enhancer positions, using as covariates the number of instances, the wild-type TF motif importance and identity, and sequence context features such as the position within the enhancer, the flanking nucleotides, and the presence at close or distal distances to all other TF motifs. The PCC between predicted and observed motif activities is shown with the green color scale on the left. The heatmap shows the contribution of each feature (columns) for each model, colored by the FDR-corrected P-value (red or blue scale depending on positive or negative association, respectively).











