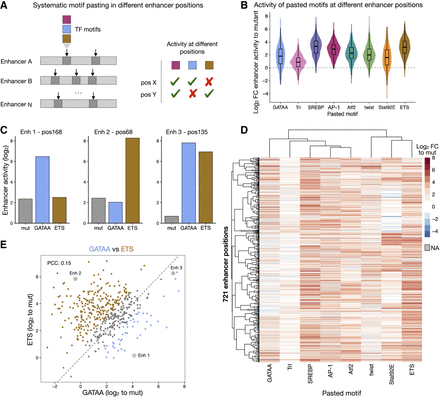
TF motifs work differently at different enhancer positions. (A) Schematics of systematic motif pasting in different enhancer positions. Eight TF motifs that showed distinct position-dependent preferences were selected and their optimal sequence was pasted in 763 positions distributed among 496 enhancers, representing different contexts. The enhancer activity of each variant was measured by STARR-seq in Drosophila S2 cells to quantify the activity of motifs at the different positions. (B) Distribution of enhancer activity changes (log2 FC to mutated sequence) across all enhancer positions for each pasted TF motif. (C) Bar plots with activity (log2) of variants of three different enhancers with a mutated sequence (gray), a GATA (blue), or a ETS (brown) motif pasted at the same position. (D) Heatmap of enhancer activity changes (log2 FC to mutated sequence) after pasting each of the eight selected TF motifs in 721 enhancer positions (positions with data for at least six motifs). TF motifs and positions were clustered using hierarchical clustering and the activity is colored in shades of red (activating) and blue (repressing); missing values are colored in gray. (E) GATA and ETS motifs work differently at different enhancer positions. Comparison between enhancer activity changes (log2 FC to mutated sequence) after pasting GATA (x-axis) or ETS (y-axis) across all enhancer positions. Positions with stronger activity of GATA or ETS (≥twofold with respect to the other motif) are colored in blue and brown, respectively. Enhancer positions shown in C are highlighted. PCC: Pearson correlation coefficient.











