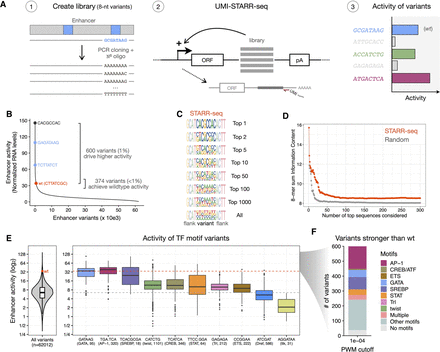
STARR-seq comprehensively assesses the activity of random variants in a specific enhancer position. (A) Schematics of STARR-seq for the analysis of random variants in an enhancer position: (1) A comprehensive library of sequence variants was generated by replacing the 8-nt stretch overlapping a GATA TF motif in the strong ced-6 enhancer with all possible 65,536 randomized nucleotides; (2) the enhancer activity of each variant was measured by STARR-seq in Drosophila S2 cells; (3) expected outcomes include the wild-type sequence (wt, blue), inactive variants (gray), and variants that recover the wild-type activity (green) or are even stronger (purple). (B) Most sequence variants exhibit low activity levels. The distribution of enhancer activity for each of the 62,012 enhancer variants with confident activity is shown. The wild-type (wt, red) sequence, the strongest GATA variant in each orientation (blue), and the strongest sequence variant are highlighted, together with the number of variants that achieve similar activity to wild type (±10%) or drive even higher activity. (C) Strong sequence variants are highly diverse. Logos with nucleotide frequency of the most-active variants in STARR-seq (1, 2, 5, 10, 50, 100, 1000, and all) and flanking nucleotides. Please note that because variants are aligned this will smear out motifs that occur at different positions. Motif finding with HOMER for these variants is shown in Supplemental Figure S2. (D) Sum of information content within the most-active 8-mers in STARR-seq (red) compared with the same after randomly sorting the variants (gray), considering different number of top sequences. (E) Distribution of enhancer activity for all 62,012 enhancer variants (left) or variants creating each TF motif (right). The activity of the wild-type sequence (wt, red dot and dashed line) or median of all variants (gray dashed line) are shown. The string of each TF motif used for the motif matching and the number of variants matching to each motif are described in the x-axis in the format “motif string (TF motif name, number of variants).” (F) Number of variants among the 600 stronger than wild type that match to motifs enriched in S2 developmental enhancers (PWM P-value cutoff 1×10−4).











