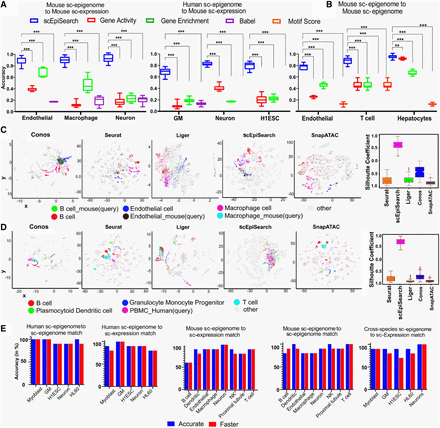
Evaluation of the accuracy of scEpiSearch. (A) Comparison of scEpiSearch with three approaches based on correlation of gene scores (activity, enrichment and BABEL-based predicted expression) for matching single-cell open-chromatin profiles to a pool of reference single-cell transcriptome. Here the reference data set consisted of single-cell expression profiles of 10,100 cells chosen from the mouse cell atlas (MCA). Accuracy here shows the percentage of query cells which had the correct cell type among the top five matches. (B) Comparison of five methods for matching query single cell open-chromatin profile to reference sci-ATAC-seq profile of ∼81,000 mouse cells published by Cusanovich et al. (2018). (***) P-value <0.001; (**) P-value <0.01. (C) Comparison of scEpiSearch integrative method using reference single-cell expression profiles of 10,100 cells from MCA data set. Here query consisted of scATAC-seq profiles of three types of mouse cells, namely, B cells, macrophages, and endothelial. The silhouette index of query cells, for being in proximity to correct reference cell types, is shown for different methods, on the right panel. (D) Evaluation of cross-species search for integrative methods and approach of scEpiSearch using human PBMC scATAC-seq profiles as query and reference single-cell expression profiles from MCA. Silhouette coefficients for human PBMCs are also shown for different methods. Here immune cells in references and query cells were considered to belong to one class, whereas other cell types as second class for calculation of silhouette coefficients. (E) Accuracy achieved by scEpiSearch engine for matching query scATAC-seq read-count matrices to its own collection of reference single-cell profiles; shown from left to right as such: (i) query human single-cell epigenome (open-chromatin) profile to reference human single-cell epigenome; (ii) query human single-cell open-chromatin profile to reference human single-cell expression collection; (iii) query mouse single-cell epigenome to reference single-cell expression profiles; (iv) query mouse single-cell epigenome (open-chromatin) to reference mouse single-cell expression profile; (v) cross-species search, query human single-cell epigenome to reference mouse single-cell expression. The y-axis shows accuracy in the percentage of query cells for which correct annotation came among the top five hits. It shows accuracies as bar plots for faster and accurate modes of scEpiSearch.











