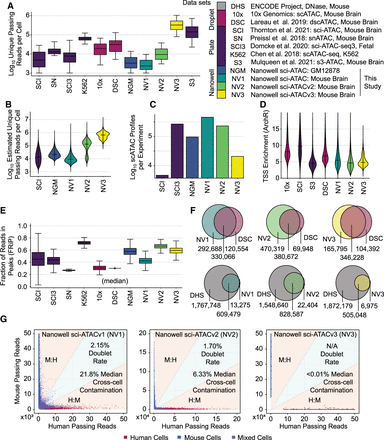
Nanowell-based sciATAC-seq quality metrics. (A) Log10 unique, passing reads per cell for nanowell sciATAC experiments compared to other single-cell ATAC-seq data sets. Data set legend on right. Data from Thornton et al. 2021 are from the nonspatial control experiment. (B) Log10 projected unique passing reads at 100% sequencing saturation (i.e., total possible unique molecules). (C) Log10 cell counts produced per preparation. (D) TSS Enrichment as determined by ArchR for single-cell ATAC-seq data sets. (E) Fraction of reads in called peaks (FRiP) for single-cell ATAC-seq data sets. (F) Called peak overlap between mouse brain data sets from this study and dscATAC (top) as well as for annotated regulatory elements in mouse (bottom). (G) Human and mouse aligned read comparisons to assess doublet rates and cross-cell contamination rates for nanowell sciATAC.











