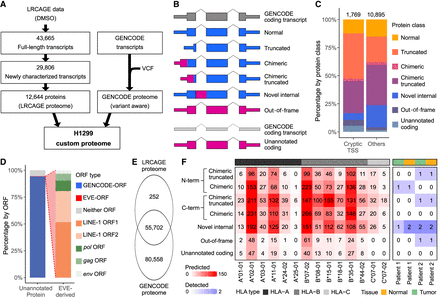
LRCAGE proteome enables the detection of unannotated proteins and noncanonical antigens in H1299 cells. (A) Flowchart of preparing H1299 custom proteome using LRCAGE data in H1299 cells. (B) Classification scheme of proteins. Pink indicates unannotated peptide sequences; blue, peptide sequences identical to GENCODE proteins. (C) Proportions of proteins in the LRCAGE proteome by protein class. (D) Percentages of unannotated proteins by their ORFs relative to GENCODE and EVE-ORFs. (GENCODE-ORF) Unannotated proteins with truncated, chimeric, chimeric truncated, or novel internal class (Fig. 1B), (EVE-ORF) unannotated proteins with out-of-frame or unannotated coding class and overlapping EVE-ORFs; and (neither ORF) unannotated proteins overlapping neither ORF type. (E) Venn diagram of peptides in H1299 whole-cell lysate MS data by proteome databases. (F) Heatmap of noncanonical antigens predicted for 14 HLA alleles (left) and antigens from H1299 HLA-pulldown LC-MS/MS data of two lung cancer patients (right). C*04:01 has no antigens predicted and was omitted.











