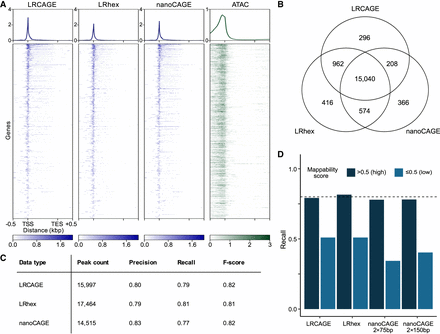
Benchmark of long-read CAGE data for promoter identification. (A) Heatmap of coverage across gene bodies. For coverage, 5′ ends of read were used for LRCAGE, LRhex, and nanoCAGE, and Tn5 insertion sites were used for ATAC-seq data. (TES) Transcription end site. (B) Venn diagram showing intersections of active GTSSs detected by LRCAGE, LRhex, and nanoCAGE (2 × 75-bp) peaks. (C) Peak count, precision, recall, and F-score by LRCAGE, LRhex, and nanoCAGE (2 × 75 bp) based on active GTSSs. (D) Recall as a function of mappability scores using 20,649 and 300 active GTSSs with high and low mappability scores, respectively. LRCAGE: 79% (16,353, high), 51% (153, low); LRhex: 82% (16,839, high), 51% (153, low); nanoCAGE 2 × 75 bp: 78% (16,085, high), 34% (103, low); and nanoCAGE 2 × 150 bp: 78% (16,122, high), 40% (121, low).











