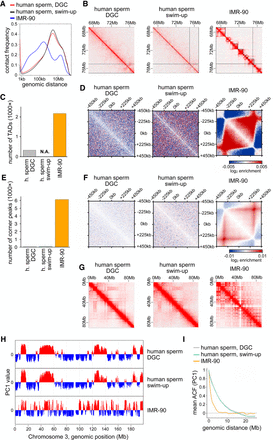
The human sperm genome is organized without TADs and corner peaks. (A) Contact frequency as a function of genomic distance for human sperm enriched by DGC and swim-up compared with human IMR-90 cells by Rao et al. (2014). (B) Normalized Hi-C matrices for the same samples as in A in the region of Chr3:67–77 Mb at 25-kb resolution with TAD calling indicated using black lines. (C) Number of TADs called for the same samples as in A. (N.A.) Artifacts in TAD calling owing to low sequencing depth. (D) Aggregate contact frequencies (coverage and distance corrected) around the 138 550- to 650-kb-long TADs called in IMR-90 cells, for the same samples as in A. (E) Number of corner peaks called for the same samples as in A. (F) Aggregate contact frequencies (coverage and distance corrected) around the 249 550- to 650-kb-long corner peaks called in IMR-90 cells, for the same samples as in A. (G) Normalized Hi-C matrices for the same samples as in A in the region of Chr3:0–90 Mb at 250-kb resolution. (H) Compartment tracks from principal component analysis for the same samples as in A. (I) The autocorrelation of the PC1 value as a function of genomic distance, for the same samples as in A.











