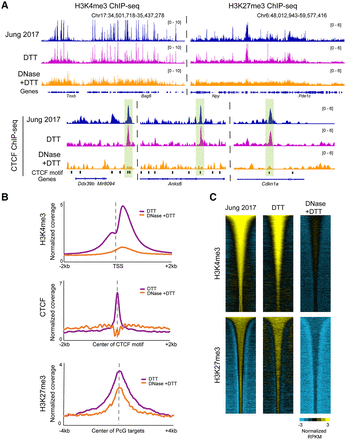
Histone modification profiling in DNase-treated sperm reveals a loss of specific signal. (A) Browser tracks for H3K4me3, H3K27me3, and CTCF ChIP-seq in sperm. In all three cases, the top panels show data from published sperm data sets (Jung et al. 2017), along with our data from DTT-treated and DNase + DTT-treated sperm underneath. (B) Metagenes aligned over the relevant peak locations (TSS, CTCF motif, and Polycomb-group [PcG] targets), showing H3K4me3, H3K27me3, and CTCF enrichment in our DTT-only and DTT + DNase data sets. (C) Heatmaps show H3K4me3 and K27me3 enrichment over all peaks from Jung et al. (2017), with data shown for Jung et al. alongside our DTT and DTT + DNase data sets.











