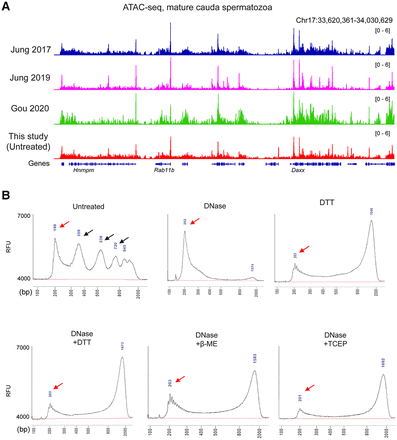
ATAC-seq profiles from sperm following DNase and DTT treatment. (A) Genome browser tracks for ATAC-seq data from untreated sperm (this study) and from three published ATAC-seq data sets for sperm (Jung et al. 2017, 2019; Gou et al. 2020). (B) Bioanalyzer traces of ATAC-seq sequencing libraries prepared from sperm samples pretreated with the indicated conditions before Tn5 transposition. Red arrows show ∼75-bp inserts (after accounting for sequencing adaptors) corresponding to open chromatin regions, and black arrows in “untreated” libraries show insert lengths corresponding to mono-, di-, and trinucleosome length inserts. Notable in all the DTT-treated libraries is a peak at ∼1.3 kb. This peak reflects a landscape of random insertions across the sperm genome, with the size of the insert being an artifact of the PCR conditions used in library preparation. We confirmed this by building a second set of ATAC-seq libraries in which the PCR extension time was extended to 2 min compared with 1 min in the libraries shown. Under these conditions, we find a ∼2.5-kb peak, confirming that PCR extension time is responsible for this peak location. (RFU) Relative fluorescence units.











