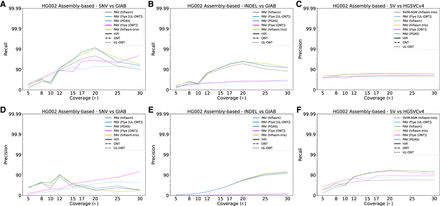
Precision and recall for variant classes as a function of LRS coverage using assembly-based algorithms for HG002. (A) Recall for HG002 for GIAB truth sets plotted against sequencing coverage for assembly-based callers across all algorithms capable of calling SNVs. (B) Recall for HG002 against HGSVC truth sets plotted against sequencing coverage for assembly-based callers across all algorithms capable of calling indels. Recall in ONT assemblies performs better at low coverages before being surpassed by HiFi assemblies at 12×. (C) Recall for HG002 against the HGSVC Freeze 4 truth set plotted against sequencing coverage for assembly-based callers across all algorithms capable of calling SVs. (D) Precision for HG002 against HGSVC truth sets plotted against sequencing coverage for read-based callers across all algorithms capable of calling SNVs. ONT methods are comparable to HiFi precision at high coverages though are noticeably worse at coverages below 15×. (E) Precision plotted against sequencing coverage for assembly-based callers across all algorithms capable of calling indels. Like read-based methods, values for all technologies and coverages remain low, likely because of the incomplete nature of indels in complex regions in the GIAB truth set. (F) Precision plotted against sequencing coverage for assembly-based callers across all algorithms capable of calling SVs.











