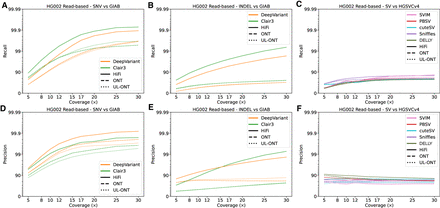
Precision and recall for variant classes as a function of long-read sequencing (LRS) coverage using read-based algorithms for HG002. (A) Recall of genome sample HG002 against Genome in a Bottle (GIAB) truth sets plotted against sequencing coverage for read-based callers Clair3 and DeepVariant. Clair3 with PacBio HiFi reaches the earliest recall plateau, whereas all callers show saturation by 20×. (B) Recall against GIAB truth sets plotted against sequencing coverage for read-based callers across all algorithms capable of calling indels. Recall of both Clair3 and DeepVariant HiFi sets outperform their ONT counterparts. (C) Recall against HGSVC truth sets plotted against sequencing coverage for read-based callers across all algorithms capable of calling structural variants (SVs). (D) Precision as a function of sequence coverage. Single-nucleotide variant (SNV) precision remains flat beyond 10×, demonstrating the ability of callers to distinguish sequencing error from true SNVs. (E) Precision plotted against sequencing coverage for read-based callers across all algorithms capable of calling indels. Precision values for all technologies and coverages remain flat, but here the increased precision of ONT callers is shown. (F) Precision plotted against sequencing coverage for read-based callers across all algorithms capable of calling SVs.











