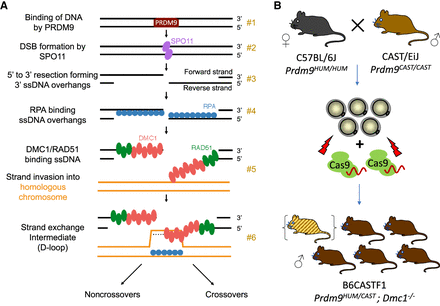
Methodology of mapping recombination intermediates using Dmc1 knockout hybrid founders. (A) Key steps in mammalian meiotic recombination. PRDM9 binds DNA at particular sequence motifs (#1), a subset of which become sites of programmed DSBs by SPO11 (#2). Strand resection creates ssDNA overhangs, which are bound by RPA first (#3) and then by DMC1 and RAD51 (#4). Binding occurs to the left of the break on the forward strand and to the right of the break on the reverse strand. A segment of DMC1-bound DNA pairs with a strand on the homologous chromosome, forming a D-loop intermediate (#5, #6). (B) Production scheme of hybrid B6CASTF1 Dmc1−/− mice. The top panel shows the parental cross, as well as the harvesting and electroporation of the F1 zygotes with CRISPR-Cas9 ribonucleoprotein targeting the Dmc1 gene. Offspring are putative biallelic knockout founders. DMC1 immunohistochemical analysis allows founders with mosaic or remnant DMC1 to be excluded from the analysis (represented as a hybrid founder mouse in parenthesis).











