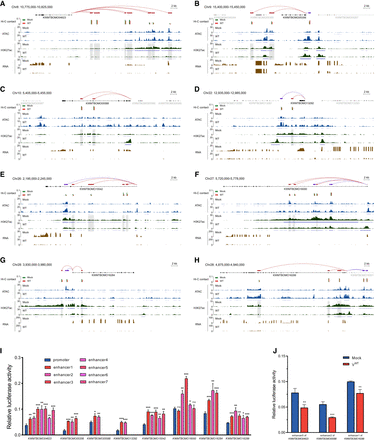
Impact of BmNPV on enhancer contacts impedes the transcriptional activation of several antiviral genes. (A–H) Normalized ATAC-seq, H3K27ac ChIP-seq, RNA-seq profiles, and Hi-C contacts showing the regulatory network of enhancers at eight antiviral gene—KWMTBOMO04623 (A), KWMTBOMO05356 (B), KWMTBOMO05588 (C), KWMTBOMO13292 (D), KWMTBOMO15542 (E), KWMTBOMO16000 (F), KWMTBOMO16284 (G), and KWMTBOMO16299 (H)—loci in uninfected cells (mock) and after 48 h of BmNPV infection (WT). Shaded regions represent decreased H3K27ac levels at the accessible enhancer regions after BmNPV infection. (I) Enhancer activity determined by dual luciferase reporter assay. Two technical replicates for each of the three biological replicates per treatment were analyzed (n = 3). The vector with null enhancer served as the control. Error bar indicates the SD. Two-tailed t-test: (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 versus the control. (J) BmNPV infection impaired the activating activity of enhancers associated with antiviral genes. Two technical replicates for each of the three biological replicates per treatment were analyzed (n = 3). Error bar indicates the SD. Two-tailed t-test: (**) P < 0.01, (***) P < 0.001 versus mock.











