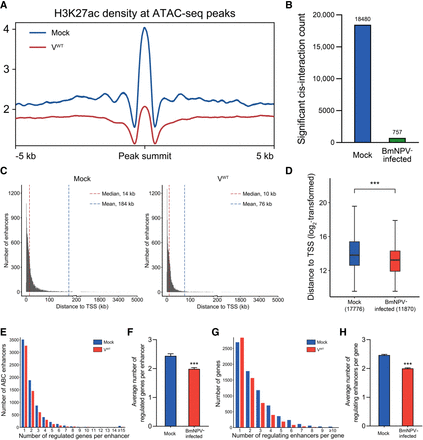
BmNPV remodels enhancer contacts by desynchronizing chromatin accessibility with H3K27ac modification and weakened cis-interaction. (A) Changes of the H3K27ac density at the ATAC-seq peaks after BmNPV infection. (B) BmNPV infection drastically attenuates cis-interaction. (C) Distances from all the predicted enhancers to the TSS of their target genes in mock- and BmNPV-infected cells. (D) Box plots of the distances showed in C. The median (center of box), lower and upper quartiles (bounds of box), and minimum and maximum values (bars) were indicated. Two-tailed Wilcoxon rank-sum test: (***) P < 10−15 versus mock. (E) Distribution histogram of the number of genes regulated by enhancer. All the identified enhancers (7269 in mock-infected cells and 5957 in BmNPV-infected cells) were analyzed. (F) Average number of genes regulated by enhancer plotted in E. Error bar indicates the SE. Two-tailed t-test: (***) P < 10−8 versus mock. (G) Distribution histogram of the number of regulating enhancers per gene in mock- and BmNPV-infected cells. All the enhancer-associated genes (7199 in mock-infected cells and 5920 in BmNPV-infected cells) were analyzed. (H) Average number of regulating enhancers per gene plotted in G. Error bar indicates the SE. Two-tailed t-test: (***) P < 10−15 versus mock.











