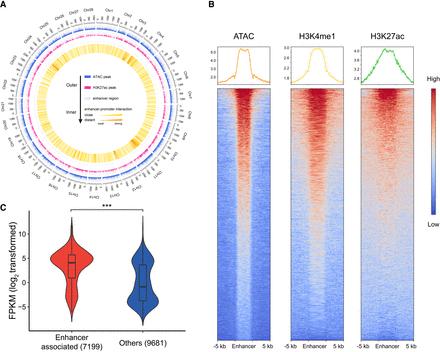
Construction of the genome-wide enhancer map. (A) Landscape of enhancer–promoter contacts and the peak distribution of chromatin accessibility and H3K27ac in the genome. (B) Enrichment of chromatin accessibility and enhancer marks around all the 7269 enhancer regions. (C) Relative expression level of enhancer-associated genes and nonassociated genes (log-transformed data). Boxes represents lower quartile, median, and upper quartile; whiskers denote the fifth and 95th percentiles. Violin plots denote the density estimates. Two-tailed Wilcoxon rank-sum test: (***) P < 10−15.











