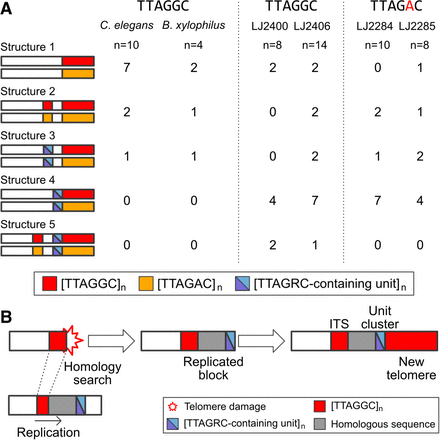
Subtelomere structures and a proposed model to explain clustered blocks of TRMs or TTAGRC-containing unit clusters in the subtelomeric regions. (A) Schematic representation of subtelomere structures in the contigs/scaffolds listed in Supplemental Table S10. We categorized the structure of subtelomeric regions (up to 200 kb from the end of the telomeric contig/scaffold) using the following criteria: (1) whether it has ITSs (shorter red blocks) and/or TTAGRC-containing unit clusters (half–sky blue/half-violet blocks) and (2) whether ITSs, unit clusters, and telomeric regions were directly attached or separated by other sequences (white, empty blocks). Each horizontal bar represents a subtelomere structure with only a telomeric TTAGRC-containing unit cluster (shown in structures 4 and 5) and an ITS or a TTAGRC-containing unit cluster closest to the telomere (shown in structures 2 or 3). Each value refers to the number of contigs/scaffolds with the corresponding structure type in each species/isolate. TTAGGC-telomere species and isolates had ITSs that were composed of TTAGGC, rather than TTAGAC, and TTAGAC-telomere species and isolates had only TTAGAC-type ITSs, too. (B) A proposed model for generating subtelomere structures responsible for structure 5. After telomere damage, HDR mechanisms can exploit homology between shortened telomeric repeats and ITSs at other loci to repair the damaged telomere. A HDR mechanism, break-induced replication (BIR), may replicate sequences near the ITS, creating a new homologous block of the original sequence block. If the original template block shows a TTAGRC-containing unit cluster, the cluster would also be replicated. Finally, a new telomere composed of TTAGGC repeats can be replenished by active telomerase.











