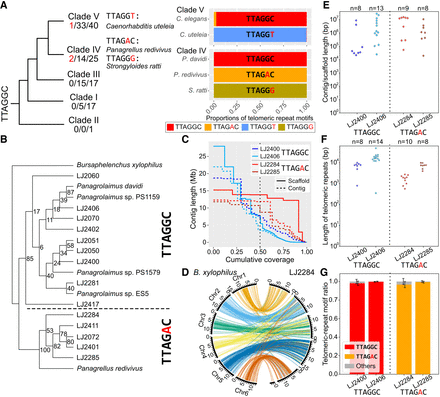
Putative novel TRMs in the phylum Nematoda and validation of the emergence of TTAGAC in the family Panagrolaimidae. (A) The cladogram was adapted and modified from Smythe et al. (2019) with permission from BMC Ecology and Evolution 2019, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). The three numbers separated by “/” under each clade indicate the number of species with novel TRMs/the number of species whose TRM was identified in our analysis/total number of species we analyzed, respectively. Changes from TTAGGC, the canonical Nematoda TRM, to the novel TRMs are highlighted in red. Below each novel TRM is a species name with the corresponding TRM. Each bar indicates the proportion of the canonical Nematoda TRM (TTAGGC) and novel TRMs in each species. The upper bar of each pair represents a control species that harbors the canonical TRM. The TRMs are represented as follows: red, TTAGGC; orange, TTAGAC; blue, TTAGGT; and khaki, TTAGGG. (B) Phylogenetic relationships of 19 Panagrolaimidae species/isolates based on their 18S rDNA sequences and their putative TRMs. Bursaphelenchus xylophilus was used as an outgroup species. The number on each node represents the bootstrap support value. (C) Contig/scaffold length distributions of the genome assemblies. The vertical dotted line indicates N50 contig/scaffold size. (D) Synteny plot of LJ2284 compared to B. xylophilus. Each colored line represents a BUSCO gene shared among the genome assemblies of our four isolates and B. xylophilus. Orange lines indicate that corresponding BUSCO genes are in Chromosome 1 in B. xylophilus. Sky blue, bluish green, yellow, blue, and vermillion represent Chromosomes 2, 3, 4, 5, and 6, respectively. We used Wong's color palette designed for color-blind individuals (Wong 2011). (E–G) The vertical dotted line separates the TTAGGC-telomere isolates (left) and the TTAGAC-telomere isolates (right). (E) Length distributions of the contigs/scaffolds containing highly clustered telomeric repeats at the end. Each dot represents each contig/scaffold, and the total number of contigs/scaffolds of each isolate (n) is indicated at the top of the graph. (F) Estimated length distributions of clustered telomeric repeats at the end of the contigs/scaffolds. Each dot indicates a telomeric-repeat cluster for each contig/scaffold, and the total number of telomeric-repeat clusters for each isolate (n) is indicated at the top of the graph. (G) The proportion of TRM types in clustered telomeric repeats. Error bars represent the SD for all clustered telomeric repeats at the end of contigs/scaffolds in each isolate.











