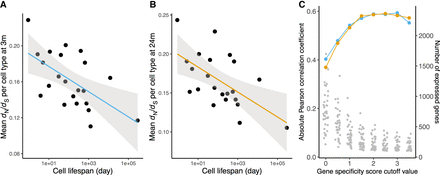
Cell life span predicts evolutionary constraint among expressed genes. The relaxation of evolutionary constraint (dN/dS) of genes expressed in 22 cell types in young (A) and old (B) mice is negatively correlated with cell life span (Pearson's r = −0.588, P = 3.9 × 10−3 and Pearson's r = −0.586, P = 4.1 × 10−3 for young and old mice, respectively). The median dN/dS is measured per cell, using cell type–specific genes (median dN/dS of genes above specificity score > 2, n = 116 to 971 genes; see Methods). The average dN/dS of a cell type is the mean of dN/dS across cells (n = 56 to 2251 cells; see Methods). The line in each figure shows an ordinary least square regression with shading of 95% confidence intervals. (C) Correlation between dN/dS and cell life span for transcriptomes of increasing cell type specificity (see Methods). The blue or yellow dots corresponding to the left axis show the correlation strength. The gray dots correspond to the right axis and show the number of expressed genes per cell type at each level of specificity.











