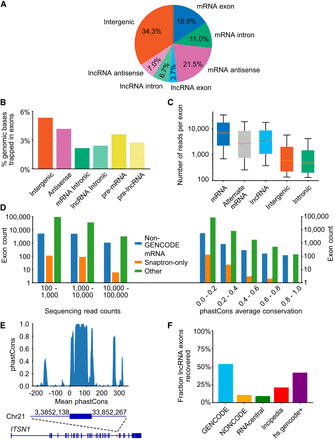
Trapped exons found in different categories of genomic region. (A) Pie chart depicting the proportions of exons from different genomic regions. The percentage of exons for indicated genomic regions relative to the total read count is indicated. (B) Bar plots showing the percent of genomic bases in a trapped exon for different categories of genomic region. (C) Boxplot depicting trapped exon sequencing read counts for different genomic regions. Whiskers depict 10th and 90th percentiles. (D) Bar plots depicting the exon counts at different read count bins (left) or average phastCons score bins (right) for different non-GENCODE v37 exon annotations. Exon counts are shown with logarithmic scale. Non-GENCODE v37 exon annotations, Snaptron database, and exons found by this exon trapping study are displayed. Average phastCons scores were calculated using the sequence of the exons. (E) Bar plot depicting phastCons (30-way) scores for +/− 200 bp around an unannotated sense intronic exon found by exon trapping in gene ITSN1. (F) Proportion of annotated exons recovered from various databases. Blue bar indicates trapped exons that are annotated in GENCODE mRNA/lncRNAs. Trapped exons annotated in other lncRNA databases are shown, with annotated GENCODE lncRNAs removed.











