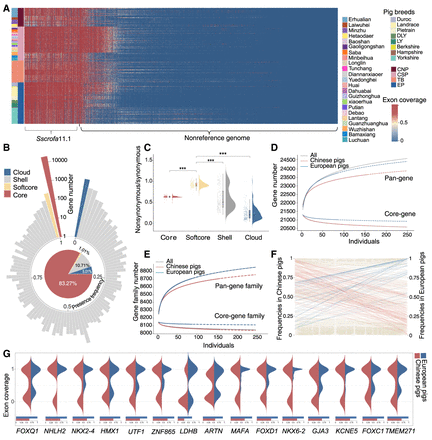
Characterization of gene PAVs. (A) The heatmap illustrates the exon coverage of dispensable genes identified in the reference genome S. scrofa 11.1 and nonreference sequences. The colors listed on the left side of the heatmaps represent different pig breeds. (B) The circle histogram depicts the distribution of gene counts with different presence frequencies in the pig population. The pie chart shows the proportion of core, softcore, shell, and cloud genes in the pig genic pangenome. (C) The ratio of nonsynonymous SNPs to synonymous SNPs in core, softcore, shell, and cloud genes. Significance values were calculated: (***) P < 1 × 10−6, Wilcoxon test. (D) Saturation curve modeling the genic pangenome and core-genome size in Chinese pigs, European pigs, and 250 individuals. The solid curve denotes fitting to the maximum gene number of sampled individuals, and the dashed curve depicts the extrapolation of the fitting. (E) Modeling of gene families in the pangenome and core genome. (F) The left y-axis and the right y-axis are occurrence frequencies of dispensable genes in Chinese pigs and European pigs, respectively. Pink and blue lines denote the genes that are favorable for Chinese pigs and European pigs, respectively. Light brown lines denote the genes with a change of presence frequency that is less than 0.4 between the two populations. (G) Comparison of probability distribution for exon coverage among populations of Chinese and European pigs for 15 genes having higher presence frequency in European pigs. The blue and red bars denote the presence frequencies of the dispensable genes in Chinese pigs and European pigs, respectively.











