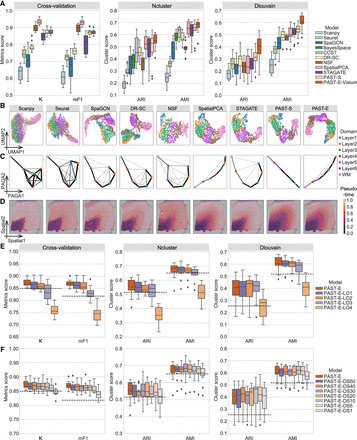
PAST robustly obtains informative low-dimensional embeddings and facilitates diverse downstream analyses. (A) Quantitative performance evaluation of spatial domain characterization on 12 DLPFC slices via supervised cross-validation and unsupervised spatial clustering with a specified number of clusters (Ncluster) and with default resolution (Dlouvain). The cross-validation performance was evaluated by the average score of Cohen's kappa value (κ) and mean F1 score (mF1), respectively, in the fivefold experiments. The spatial clustering performance was evaluated by adjusted rand index (ARI) and adjusted mutual information (AMI). The center line, box limits and whiskers in the boxplots are the median, upper and lower quartiles, and 1.5× interquartile range, respectively. (B) UMAP visualization, (C) PAGA trajectory inference results, and (D) DPT pseudotime analysis results of Slice 151,673. (E) The number of shared spatial domains in external reference data gradually deceased. (F) The spots in external reference data of PAST were down sampled with a stratified random sampling strategy to construct reference data with various scales. The horizontal dashed lines represent the corresponding median scores of STAGATE.











