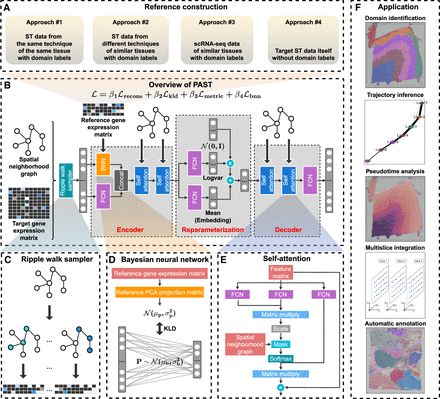
The PAST framework. (A) Four approaches for PAST to construct reference data, including three for external reference data (PAST-E) and one from the target data itself (PAST-S). (B) PAST is built on a variational graph convolutional autoencoder. The encoder of PAST consists of three layers, including the concatenation of a BNN and a FCN as the first layer and two self-attention modules as the subsequent two layers. The reparameterization module acquires the mean and log-variance matrix based on two FCNs, in which the mean matrix is the latent embedding obtained of PAST. The decoder of PAST is also a three-layer network, including a FCN layer and two stacked self-attention modules. The loss function of PAST consists of four parts, including reconstruction loss Lrecons, Kullback-Leibler divergence (KLD) loss Lkld, metric learning loss Lmetric and loss of BNN module Lbnn. (C) Ripple walk sampler samples high-quality subgraphs based on the spatial neighborhood graph and outputs minibatch gene expression matrices. (D) BNN module integrates shared biological variation through restricting the KLD distance between prior Gaussian distribution parameterized by reference data and Gaussian distribution parameterized by parameters of BNN. (E) The self-attention mechanism captures spatial correlation between neighbor spots, where FCN is used to generate queries, keys, and values for the calculation of attention weights. (F) The latent embeddings, that is, mean matrix in the reparameterization module, obtained by PAST facilitate various tasks including domain identification, trajectory inference, pseudotime analysis, multislice integration, and automatic annotation.











