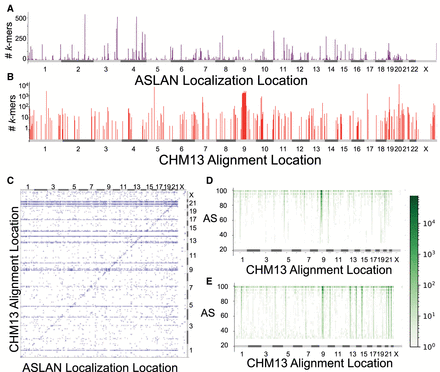
Characterizing nonconcordance between ASLAN localizations and CHM13 alignments and potential hotspots of genetic diversity. (A) Distribution of reads that failed to align to the T2T-CHM13 assembly but that were successfully localized via ASLAN. (B) Distribution of reads for which the localization region predicted by ASLAN contained the location the read aligned to on T2T-CHM13 but for which the T2T-CHM13 alignment score was less than 90. These regions may indicate new hotspots for genetic diversity. (C) Joint-plot of regions where ASLAN localization and T2T-CHM13 alignments were in disagreement with one another and where the T2T-CHM13 alignment score was less than 90. These may indicate sequences that are not represented in the T2T-CHM13 but that are somewhat homologous to a different region in T2T-CHM13 and may be mismapped. (D,E) Loci and alignment score distribution between the T2T-CHM13 alignments for k-mers with ASLAN localizations in agreement with each other (D) and in disagreement with each other (E). We see that k-mers in disagreement have significantly lower alignment scores, suggesting that imperfect alignments to T2T-CHM13 may actually be originating from a human genome sequence not well represented on T2T-CHM13.











