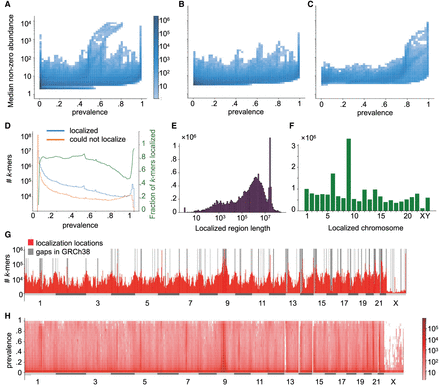
ASLAN performance on unmapped reads. (A) Distribution of prevalence and abundance (median of nonzero counts) for all 100-mers extracted from unmapped reads. (B) Distribution of prevalence and abundance for 100-mers that localized to autosomes. (C) Distribution of male prevalence and abundance for 100-mers that localized the Y Chromosome. (D) Number and fraction of 100-mers that ASLAN could and could not localize, given their prevalences across the iHART population. (E) Distribution of localized region length. (F) Number of k-mers localized to each chromosome. (G) Distribution of localization location in reference to GRCh38, with gaps annotated. (H) Distribution of k-mer localization location and prevalence.











