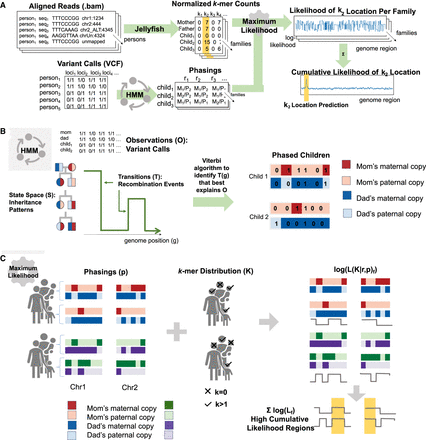
Pipeline for ASLAN and its components. (A) Overall pipeline for extracting k-mers, phasing families, and localizing k-mers based on phasings and k-mer distributions. (B) Simplified schematic of the hidden Markov model used for the phasing algorithm, in which the goal is to identify the inheritance patterns and recombination points that best explain the variant calls in a family. (C) Simplified schematic of the maximum likelihood model to identify the most likely region of a genome that a k-mer originates from, given the distribution of the k-mer and phasing patterns within and across families.











