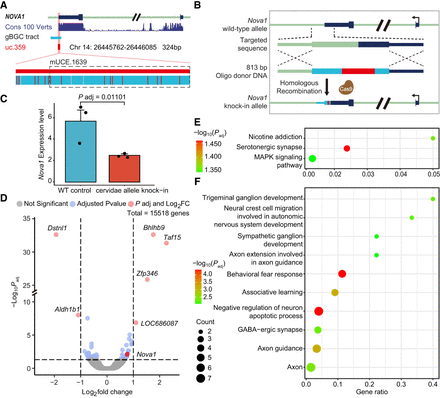
gBGC-induced functional alterations of UCEs in Cervidae. (A) uc.359 in Cervidae. Sixteen W → S cervid-specific substitutions are shown as red short lines. The black dotted rectangle is the 271-bp mUCE.1639 in our data set. (B) Gene editing abstract of the cervid-specific gBGC tract; the target region was replaced with an 811-bp cervid-specific gBGC tract. (C) Knock-in heterozygous rats showed a significant decrease of Nova1 expression. (D) Volcano plot comparing wild-type and knock-in rats. Cutoff values of |fold change| > 2 and FDR < 0.05 were used to identify differentially expressed genes. (E) KEGG pathways of 72 significantly expressed genes in D. (F) Top 10 nervous system-related GO items of 72 significantly expressed genes in D.











