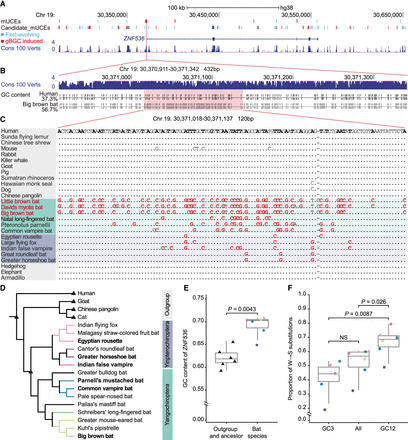
Example of gBGC-induced accelerated evolution of UCE in Chiroptera. (A) All mUCEs and candidate mUCEs in the region of ZNF536 ± 100 kb. The elements in blue indicate fast-evolving elements, whereas gBGC-induced elements are shown in red. (B) The 432-bp mUCE.2036 is located in the ZNF536 promoter region. Blue bars indicate conserved regions in 100 vertebrates. The GC content of mUCE.2036 was 37.3% and 56.7% in humans and big brown bats (Vespertilionidae), respectively. (C) Sequence alignment of a 120-bp subregion inside mUCE.2036. The shaded species names indicate species in Chiroptera, and the names in red indicate bats in Vespertilionidae. W → S substitutions are highlighted in red. Dots in the sequence alignment refer to bases that are identical to those in the human genome. (D) Phylogeny of 19 analyzed species. Four outgroup species and two ancestral nodes are marked with a black triangle, and selected bat species for subsequent analyses are marked in bold. The branches of different bat families are shown in different colors. (E) ZNF536 GC content comparison between six bat species and outgroup and ancestor. The dot colors are the same as that of the corresponding bat family in D. (F) Comparison of proportion of W → S substitutions in different codon positions.











