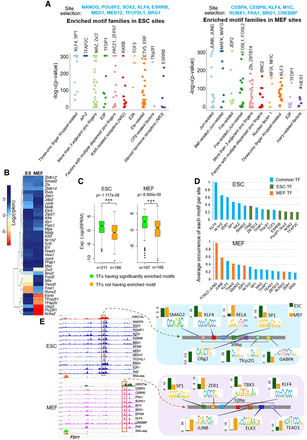
Motif compositions of enhancers in ESCs and MEFs show flexible enhancer grammar. (A) The 10 most significantly enriched TF motif families identified in the top TF-bound regulatory sites (nonoverlapping promoters) from mouse ESCs and MEF cells. For each of the 10 motif families, the motifs with the smallest P-values associated with highly expressed TFs are listed at the top. (B) The expression levels of the TFs presented in A and D, in ESCs and MEF cells. (C) The expression levels of TFs that have enriched motifs are significantly higher than those that do not have enriched motifs, with p-value = 1.117 × 10−8 in ESC and 8.565 × 10−9 in MEF (Wilcoxon test). (D) The average occurrence per site of the 15 most occurring motifs in the ESC- and MEF-specific enhancers (mid 700 bp) for common genes. (E) The TF motif composition of an ESC and an MEF enhancer linked to a commonly expressed gene, F2rl1. The expression levels (in RPKM) of each TF in the two cells (ESC as green and MEF as orange) are shown as a bar plot beside their motifs.











