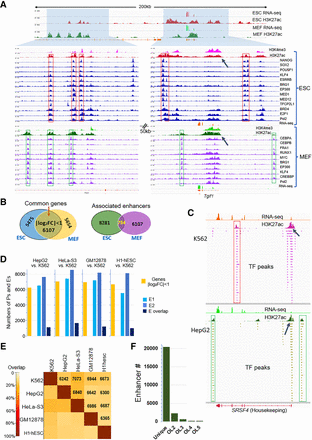
Commonly expressed genes across multiple cell types associated with cell type–specific enhancers. (A) RNA-seq and ChIP-seq signals of H3K4me3, H3K27ac, and multiple TFs around a highly expressed gene Tgif1 in mouse ESCs and MEF cells. The gene promoters are marked with arrows; enhancers, with frames. Tgif1 is the nearest active promoter to the shown enhancers. (B) Number of commonly expressed genes whose expression levels vary within twofold between ESC and MEF, and numbers of their associated strong enhancers and their overlaps between the two cells. (C) An example of cell type–specific enhancers for a housekeeping gene SRSF4 in K562 and HepG2 cells. (D) Analysis similar to what is presented in B, between K562 and another cell line. (E) Pairwise analysis of common genes (numbers in the grid) and overlap rates of associated strong enhancers among five human cell lines: K562, HepG2, GM12878, HeLa-S3, and H1-hESC. (F) For the 6272 commonly expressed genes among the five cell lines, the number of associated strong enhancers that are unique in one cell type or are shared by two, three, four, and five cell types. (OL) Overlap.











