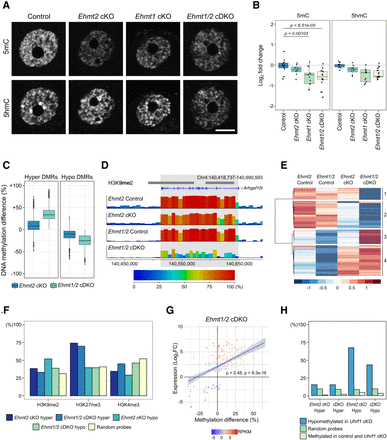
DNA methylation changes in Ehmt2 cKO and Ehmt1/2 cDKO oocytes. (A) Representative IF images of NSN oocytes showing 5mC and 5hmC in control, Ehmt2 cKO, Ehmt1 cKO, and Ehmt1/2 cDKO oocytes. (B) Boxplots of quantitation of IF images. Dots represent individual oocytes. Analysis is based on two mice per genotype and two replicate experiments for each antibody. (C) Boxplot showing the methylation difference of Ehmt1/2 cDKO DMRs (hyper and hypo) in controls versus Ehmt2 cKO and controls versus Ehmt1/2 cDKO oocytes. (D) Genome screenshot showing example of a region uniquely hypomethylated in Ehmt1/2 cDKO oocytes spanning the Arhgef10l transcript. Annotation tracks show position of H3K9me2-enriched domains and oocyte transcripts. (E) Heatmap showing clustering analysis of Ehmt1/2 cDKO hypo- and hypermethylated domains. (F) Bar chart showing percentage overlap of DMRs and random probes with H3K9me2, H3K37me3, and H3K4me3. H3K9me2: Ehmt2 cKO hyper adj. P = 0.0136, Ehmt2 cKO hypo P < 0.0001, Ehmt1/2 cDKO hypo P < 0.0001; H3K27me3: Ehmt2 cKO hyper P < 0.0001, Ehmt1/2 cDKO hyper P < 0.0001; H3K4me3: all genotypes P < 0.0001. (G) Plot showing correlation between methylation changes (% methylation difference) and expression changes (Log2FC) of DEGs in Ehmt1/2 cDKO oocytes. Relative expression levels (RPKM) of each gene are indicated by the color scale. Spearman's correlation is shown. (H) Bar chart showing percentage overlap of DMRs with 100-CpG windows that are hypomethylated in Uhrf1 cKO oocytes (>20% methylation difference) (Maenohara et al. 2017), random 100-CpG windows, and 100-CpG windows that are highly methylated (>75%) in WT and Uhrf1 cKO oocytes. Ehmt2 cKO hyper adj. P = 0.0404; Ehmt1/2 cDKO hyper, Ehmt2 cKO hypo, and Ehmt1/2 cDKO hyper P < 0.0001.











