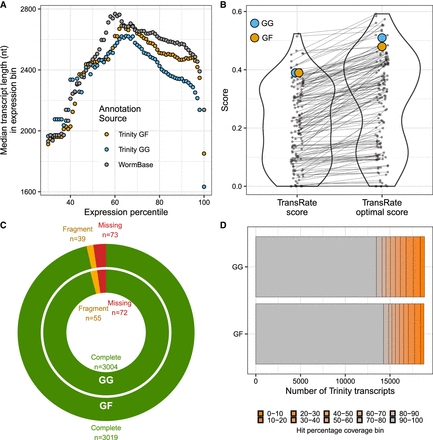
Benchmarking transcriptome assembly in C. elegans. (A) Median length of transcripts (N50) across all expression bins (ExN50) for the genome-free (GF) and genome-guided (GG) assembly compared with the WormBase annotation. (B) TransRate scores and TransRate optimal scores of the C. elegans GF and GG transcriptome assembly compared with other assemblies of different species and various assembly algorithms (Smith-Unna et al. 2016). (C) BUSCO analysis statistics of the GF and GG transcriptome assembly. (D) Bar chart summarizing transcript sequence coverage comparing GF and GG transcripts to the corresponding WormBase annotation. Completeness (hit percentage coverage) was determined by percentage overlap between the predicted transcript sequence with its equivalent WormBase annotation.











