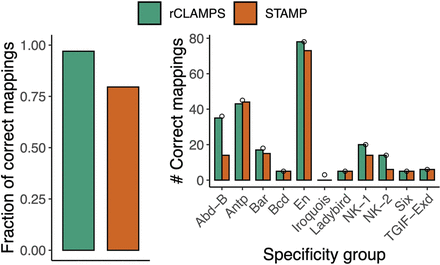
Inferred mappings to contact map are highly accurate. We compare mappings inferred by rCLAMPS (green) and computed based on direct PWM multiple alignment performed by STAMP (red) to those that are known experimentally for TFs that have identical amino acids within their base-contacting positions. (Left) The fraction of predicted mappings that is identical to the experimental mappings. (Right) The number of identical mappings when homeodomains are classified with respect to “specificity groups” as determined by Noyes et al. (2008b). Small circles represent the total number of homeodomains considered in each of these specificity groups.











