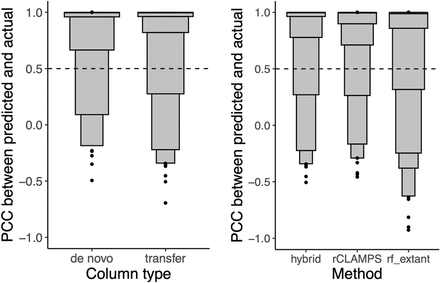
Column transfer via learned structural mappings is highly effective for predicting DNA-binding specificities for mutant TFs. (Left) For the hybrid approach, the PWM columns are binned according to whether they are predicted de novo or via transfer, and the PCCs of the predicted versus actual columns are shown in letter-value plots. Although 90% of de novo predictions are in agreement with their experimentally determined counterparts, an even higher 93% of predictions via transfer are in agreement. (Right) For each of the homeodomains that were also not part of the rf_extant training set and by considering only columns for which transfer was used by our hybrid approach, we compute the PCC between the actual specificity for a column and that predicted by our hybrid approach (hybrid), our de novo linear approach (rCLAMPS), and the rf_extant model (rf_extant). We find that 93% of transferred predictions are in agreement with their experimentally determined counterparts, compared with 91% and 85% of de novo predictions for rCLAMPS and rf_extant, respectively.











