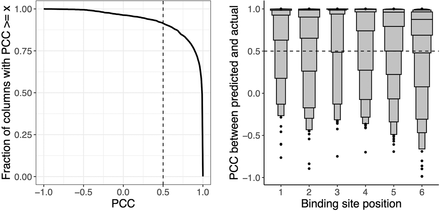
Probabilistic DNA recognition code for homeodomains derived from automatically inferred structural mappings has excellent de novo predictive performance. We compare agreement between predicted PWM columns and corresponding experimental PWM columns for 763 homeodomain proteins in a strict holdout validation setup. (Left) Considering all homeodomain binding site positions together, as different thresholds of PCC are considered (x-axis), the fraction of column pairs that have PCC greater than this threshold is plotted (y-axis). Our nominal threshold for agreement (PCC ≥ 0.5) is shown as a dashed vertical line. (Right) For each binding site position within the homeodomain contact map (x-axis), we display the PCC agreement scores (y-axis) for the paired columns at that binding site position, visualized as letter-value (or boxen) plots. In a letter-value plot, the widest box shows the value range spanned by half the data (from the 25th to 75th percentiles), whereas each successively narrower pair of boxes together show the value range spanned by half the remaining data. The PCCs at the 25th percentile for positions 1–6 are 0.98, 0.90, 1.00, 0.97, 0.79, and 0.69, respectively.











