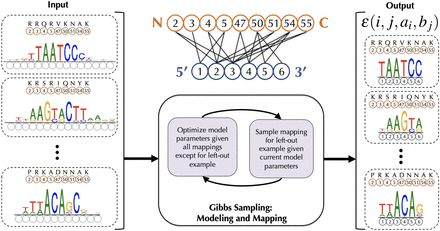
Schematic of our procedure rCLAMPS for jointly learning protein–DNA interaction interfaces and structure-aware recognition codes for TFs of the same structural family. (Middle, top) Our approach first analyzes protein–DNA co-complex structural data for a TF family to determine commonly observed pairwise contacts between positions in the protein (orange circles) and positions within DNA (blue circles) that together comprise a structural interface or “canonical” contact map. Here we show such a contact map for the homeodomain TF family, with protein positions corresponding to match states in Pfam homeodomain model PF00046 (relabeled as canonical homeodomain positions from Noyes et al. 2008b). (Left) Given a set of TFs and their corresponding DNA-binding specificities as PWMs, the positions (and amino acids) within each TF that interact with DNA are known (orange circles and amino acids above), but initially the positions within the PWMs that are contacted by these amino acids are not known (dotted blue circles). (Middle, bottom) We use a Gibbs sampling approach to map the PWM positions to DNA positions within the contact map wherein base preferences at each nucleotide position are described in terms of additive amino acid–base contact energies. (Right) After Gibbs sampling is complete, we have a mapping of each TF–PWM pair to the TF family contact map, along with a linear recognition code for the TF family that consists of pairwise energy estimates for each amino-to-base pairing in each of the (i, j) amino acid–nucleotide position pairs in the contact map.











