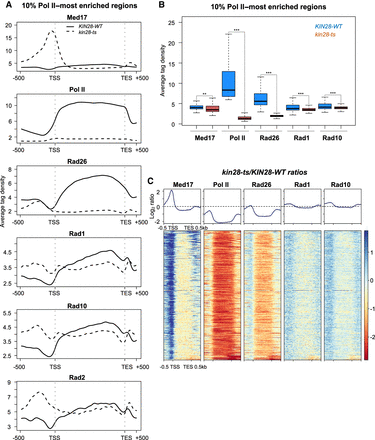
Effect of the kin28-ts mutation on genome-wide Mediator, Pol II, Rad26, Rad1, and Rad10 occupancy at transcribed regions. (A) Average tag density in Med17 (Mediator), Pol II, Rad26, Rad1, and Rad10 ChIP (from upper to lower panels) on the 10% Pol II–most enriched regions (scaled windows for 500 bp before TSS, between TSS and TES, and 500 bp after TES). For comparison, the Rad2 ChIP-seq data were from Georges et al. (2019). Average tag density in WT strains is indicated as a full line, whereas average tag density in the kin28-ts strains is indicated as a dashed line. (B) Boxplots showing changes in Med17 (Mediator), Pol II, Rad26, Rad1, and Rad10 ChIP-seq on the 10% Pol II–most enriched regions in WT (in dark blue) and the kin28-ts strains (in brown). The asterisks represent a significant difference between the WT and the mutant at P-value < 1 × 10−10 (**) or < 2.2 × 10−16 (***) in a Wilcoxon rank-sum test. (C) Heat maps of Mediator (Med17), Pol II, Rad26, Rad1, and Rad10 ChIP-seq occupancy ratios between the kin28-ts mutant and the WT on the 10% Pol II–most enriched regions (scaled windows for 500 bp before TSS, between TSS and TES, and 500 bp after TES), sorted by decreasing Mediator occupancy ratio. Average log2 ratios are shown in upper panels.











