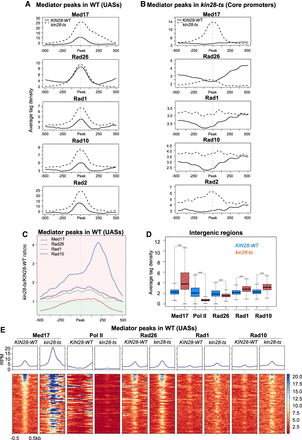
Effect of the kin28-ts mutation on genome-wide Mediator, Rad26, Rad1, and Rad10 occupancy at intergenic regions. (A) Average tag density in Med17 (Mediator), Rad26, Rad1, and Rad10 ChIP-seq (from upper to lower panels) around Med17 (Mediator) enrichment peaks (−500 bp to +500 bp) determined in WT (UASs). The Rad2 ChIP-seq data were from Georges et al. (2019). Average tag density in WT strains is indicated as a full line, whereas average tag density in the kin28-ts strains is indicated as a dashed line. (B) Average tag density in Med17 (Mediator), Rad26, Rad1, and Rad10 ChIP-seq (from upper to lower panels), around Med17 (Mediator) enrichment peaks (−500 bp to +500 bp) determined in kin28-ts (core promoters). Rad2 ChIP-seq data were used (Georges et al. 2019). Average tag density in WT strains is indicated as a full line, whereas average tag density in kin28-ts strains is indicated as a dashed line. (C) Mediator (in blue), Rad26 (in brown), Rad1 (in green), and Rad10 (in purple) occupancy ratios between the kin28-ts mutant and the WT around Med17 (Mediator) peaks (UAS, 1000-bp window). The maximum of the Rad1–Rad10 occupancy ratios is located at UAS, and the maximum for Mediator occupancy ratios is shifted to core promoters. Rad26 occupancy ratio is only slightly increased at UASs. For clarity, the background of the plot for ratios >1 is colored in clear red, and the background of the plot for ratios <1 is colored in clear green. (D) Boxplots showing changes in Med17 (Mediator), Pol II, Rad26, Rad1, and Rad10 ChIP-seq at intergenic regions (intergenic regions for Pol II–transcribed genes in tandem or in divergent orientation, excluding intergenic regions encompassing Pol III–transcribed genes, centromeres, telomeres) in the WT (in dark blue) and the kin28-ts strains (in brown). The asterisks represent a significant difference between the WT and the mutant at P-value < 2.2 × 10−16 in a Wilcoxon rank-sum test. (E) Heat maps of Mediator (Med17), Pol II, Rad26, Rad1, and Rad10 ChIP-seq profiles centered on Mediator (Med17) enrichment peaks determined in WT (UASs, −500 bp to +500 bp), sorted by decreasing Mediator occupancy. WT and kin28-ts strains were compared. Median tag density profiles in RPM are shown in upper panels.











