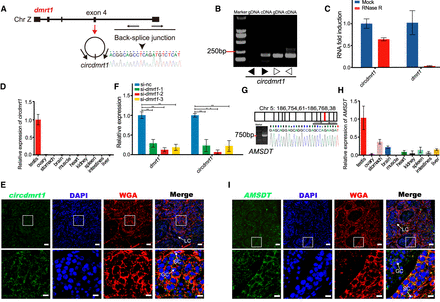
Characteristics of circdmrt1 and AMSDT in tongue sole testis. (A) The genomic locus of circdmrt1 in the dmrt1 gene. Circdmrt1 is produced at the dmrt1 gene locus containing exon 4. The back-splice junction of circdmrt1 was identified by Sanger sequencing. Arrows represent divergent primers binding to the genome region of circdmrt1. (B) RT-PCR products with divergent and convergent primers showing circularization of circdmrt1 in 6-mpf tongue sole testis. (cDNA) Complementary DNA, (gDNA) genomic DNA. The black and white arrows represent the divergent and convergent primers, respectively. (C) qRT-PCR results revealing the abundance of circdmrt1 and dmrt1 mRNA in 6-mpf tongue sole testis treated with RNase R. The amounts of circdmrt1 and dmrt1 mRNA were normalized to the values measured in the mock group. The blue and red bars represent the mock-treated or RNase R-treated group, respectively. (D) Relative quantification for circdmrt1 in 10 tissues of tongue sole. (E) RNA fluorescence in situ hybridization for circdmrt1 in tongue sole testis. Circdmrt1 probe was labeled with fluorescein amidites (FAM) and detected by TSA-FAM (green signals). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). The cell membrane was stained with the wheat-germ agglutinin (WGA)/Alexa Fluor 555 conjugate dye. The bottom row shows enlargement of the regions outlined in the top row. Scale bars in top rows are 20 μm, and bars in bottom rows are 5 μm. (F) Expression levels of dmrt1 and circdmrt1 in tongue sole testis treated with dmrt1 siRNA. The transcription levels were normalized to Actb1 levels. (G) RT-PCR analysis for AMSDT in cDNA of 6-mpf tongue sole testis. The red line in the upper panel shows the location of AMSDT in Chr 5. The lower panel represents the Sanger sequencing of RT-PCR products of AMSDT including the binding sites with cse-miR-196. (H) Relative quantification for AMSDT in 10 tissues of tongue sole. (I) RNA fluorescence in situ hybridization for AMSDT in tongue sole testis. The AMSDT probe was labeled with fluorescein amidites and detected by TSA-FAM (green signals). Nuclei were stained with DAPI. The cell membrane was stained with wheat-germ agglutinin/Alexa Fluor 555 conjugate dye. The bottom row shows enlargement of the regions outlined in the top row. Scale bars in top rows are 20 μm, and bars in bottom rows are 5 μm. Data in C, D, F, and H are the means ± SD of three experiments. (**) P < 0.01, two-tailed t-test. Abbreviations: (LC) Leydig cell, (SC) Sertoli cell, (GC) germ cell.











