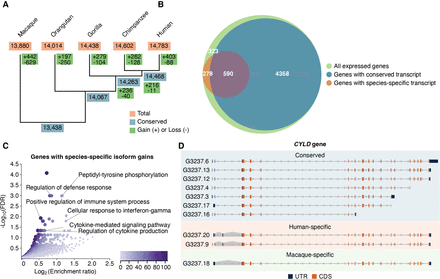
Transcript expression gains and losses in the primate lineage. (A) Dynamics of transcript expression gains and losses in five primate species (inferred by Wagner parsimony). The phylogenetic tree is built based on the known phylogeny. Transcript expression gains and losses in macaque versus great apes are shown in macaque's branch, although the direction (gain or loss in macaque or great apes) cannot be determined. (B) Overlap of genes expressing highly conserved (blue) and species-specific transcripts (orange). (C) Functional enrichment for genes displaying species-specific isoform innovations (N = 868 genes). Overrepresentation analysis (ORA) was conducted using the Gene Ontology Biological Process database (affinity propagation clustering). False-discovery rates (FDRs) were adjusted using the Benjamini–Hochberg method (Benjamini et al. 2001). The color legend indicates the number of overlapping genes between the gene set under evaluation and the pathway gene set. (D) Example of a gene, CYLD, expressing transcripts in all five species (conserved; blue shadow), exclusive expression in human (human-specific; orange shadow), and exclusive expression in rhesus macaque (macaque-specific; green shadow). Gray shadows indicate species-specific splice junctions. Untranslated regions (UTRs) and coding sequences (CDSs) are colored in blue and orange, respectively.











