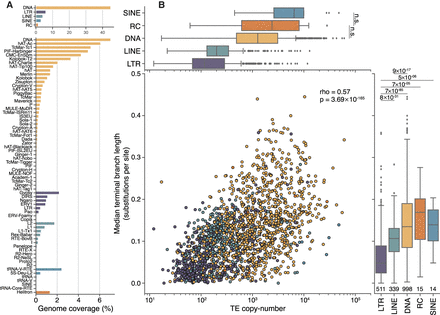
Genome proportions, copy number, and median age differ between TE classes. (A) DNA transposons, including rolling-circle elements (Helitrons), take up approximately four times more genomic space than retroelements and contain a greater number of distinct superfamilies. (B) Overall, there is a moderate correlation between the copy number of TE families and their median age (Spearman's ρ = 0.57, P = 3.69 × 10−165). LTR elements, on average, are younger than other classes (lower values on the y-axis), and DNA transposons are typically older. Numbers underneath the box plots are the number of distinct TE families used in this analysis. Significance was calculated using Wilcoxon rank-sum tests between each TE class, using a Bonferroni-corrected P-value threshold of 0.001 for determining significance. For clarity, only the two nonsignificant tests are shown in the top panel.











